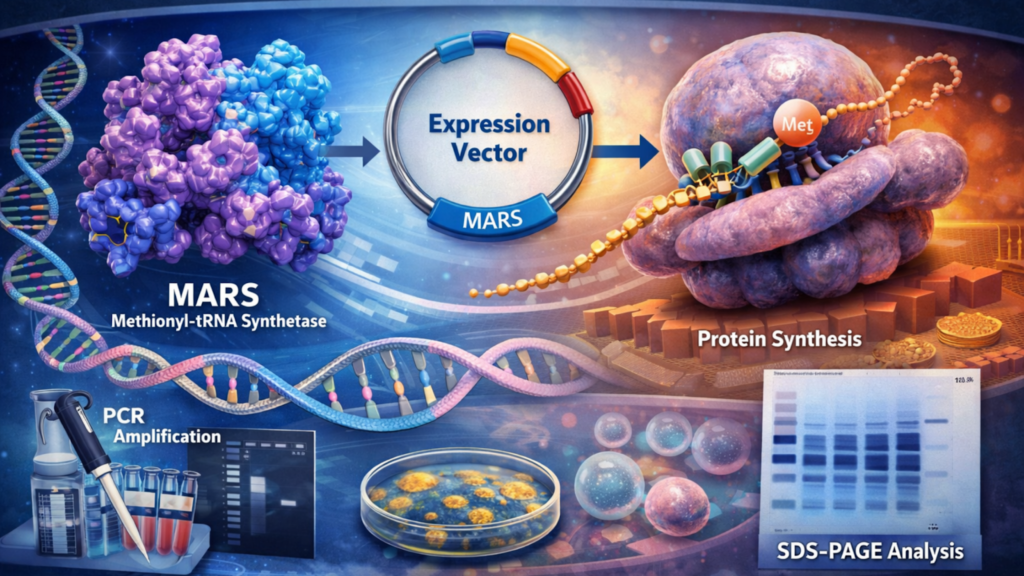
Protein synthesis is an essential process in all living cells that allows cells to produce proteins required for growth, signaling, and survival. This process depends on enzymes such as methionyl-tRNA synthetase working together in a precise manner. Also known as MARS, methionyl-tRNA synthetase attaches methionine to its corresponding tRNA during the early steps of translation. This is why it is often studied in molecular and cellular biology research to understand:
- How translation initiation is regulated.
- How protein synthesis becomes defective.
- How cells maintain protein homeostasis.
MARS cloning and expression allow researchers to produce the protein in controlled systems and study its function in detail.
Biological Significance of MARS
MARS belongs to the family of aminoacyl-tRNA synthetases. These enzymes couple amino acids to their corresponding tRNAs to ensure accurate translation. Methionine is an amino acid that initiates protein synthesis.
MARS activity has a direct influence on the initiation of translation and overall protein production. Besides its main enzymatic role, MARS also takes part in regulatory processes such as:
- How cells respond to stress.
- How protein synthesis is adjusted based on the cell’s metabolic state.
These key roles make MARS an attractive target for mechanistic studies, necessitating reliable systems for its recombinant production and analysis.
Cloning Strategy for MARS
Isolation of Coding Sequence
The corresponding coding sequence is isolated from:
- A cDNA library
- Reverse-transcribed RNA derived from human cells
This step ensures that the full protein-coding region is obtained for downstream applications.
Primer Design and Gene Amplification
Primers must be designed to amplify the full-length open reading frame of MARS. It should also remain compatible with the selected expression vector. MARS encodes an enzymatically active protein. This is why high-fidelity DNA polymerases are commonly used during PCR amplification to minimize sequence errors.
Insertion into Expression Vectors
The amplified MARS sequence is inserted into an expression vector. The selection of an expression vector depends on the host system and the intended downstream applications.
Incorporation of Affinity or Epitope Tags
Affinity tags are used to purify proteins. They also enable detection in downstream assays. Commonly used affinity tags include polyhistidine or FLAG tags.
Screening and Validation of Clones
Restriction enzyme analysis is then used to screen candidate clones. DNA sequencing confirms positive clones to verify correct gene insertion, proper orientation, and overall sequence integrity
Expression Systems for Recombinant MARS
The success of the experiment greatly depends on the expression system. Bacterial systems, particularly Escherichia coli, are used because of their simplicity, rapid growth, and cost-effectiveness.
While bacterial expression produces large quantities of recombinant MARS, it has some limitations. For example, it lacks eukaryotic post-translational modifications. There can be folding or solubility issues.
Mammalian expression systems provide a more relevant environment that enables proper folding and modification of MARS. Transient or stable expression in cultured cells allows researchers to study recombinant MARS in a context that closely resembles endogenous conditions. However, mammalian systems require more resources and yield proteins in lower quantities.
Validation of MARS Expression at the Protein Level
Verification of expression is an essential step after cloning and transformation. Protein-level validation ensures that the cloned gene is not only transcribed but also translated into a protein of the expected size and integrity. SDS-PAGE is used to analyze cell or tissue lysates.
The expected molecular weight of the detected MARS confirms successful expression. It also provides insights into the stability of the protein. An anti-MARS antibody is used to specifically identify the recombinant protein.
Challenges in Cloning and Expression of MARS
Cloning and expression of MARS can be technically demanding due to the following challenges:
- Limited solubility in bacterial systems
- Formation of inclusion bodies
- Need for optimization of expression conditions
- Requirement for refolding strategies
- Overexpression effects in mammalian cells
- Difficulty interpreting functional results
- Differences between recombinant and endogenous MARS
- Lack or alteration of post-translational modifications
- Loss or change of normal protein–protein interactions
- Risk of experimental artifacts caused by overexpression
- Need for proper controls and comparative analyses
Applications of Recombinant MARS
| Application | Description |
| Functional enzymatic assays |
|
| Protein–protein interaction studies |
|
| Structural biology and mechanistic research |
|
| Assay development and translational studies |
|